Lessons in Antisense
Lesson 17 – The Science of ASO Design and Screening Using the FDA Guidance for Nano-rare Patients
August 11, 2025 by Dr. Stan Crooke

Introduction
Though ASO design and screening are much more rational, rapid and efficient than traditional drug discovery and continues to evolve as we learn more, screening appropriate numbers of PS ASOs through several steps is still essential. Since the FDA guidance for ASOs for nano-rare patients supports treating patients with only in vitro pharmacological data and a single GLP three-month toxicology study, thorough design and screening are especially critical. In this lesson, I plan to explain the basis of the n-Lorem ASO design and screening process, the reasons for each step, each analyte that is used, and the minimum number of ASOs studied at each step. Because the screening for ASOs to be used to treat CNS diseases is more cumbersome and involved than that used for targets in other organs, I will focus on the design and screening of PS ASOs designed to activate RNase H1 and degrade target RNAs for targets in the CNS.
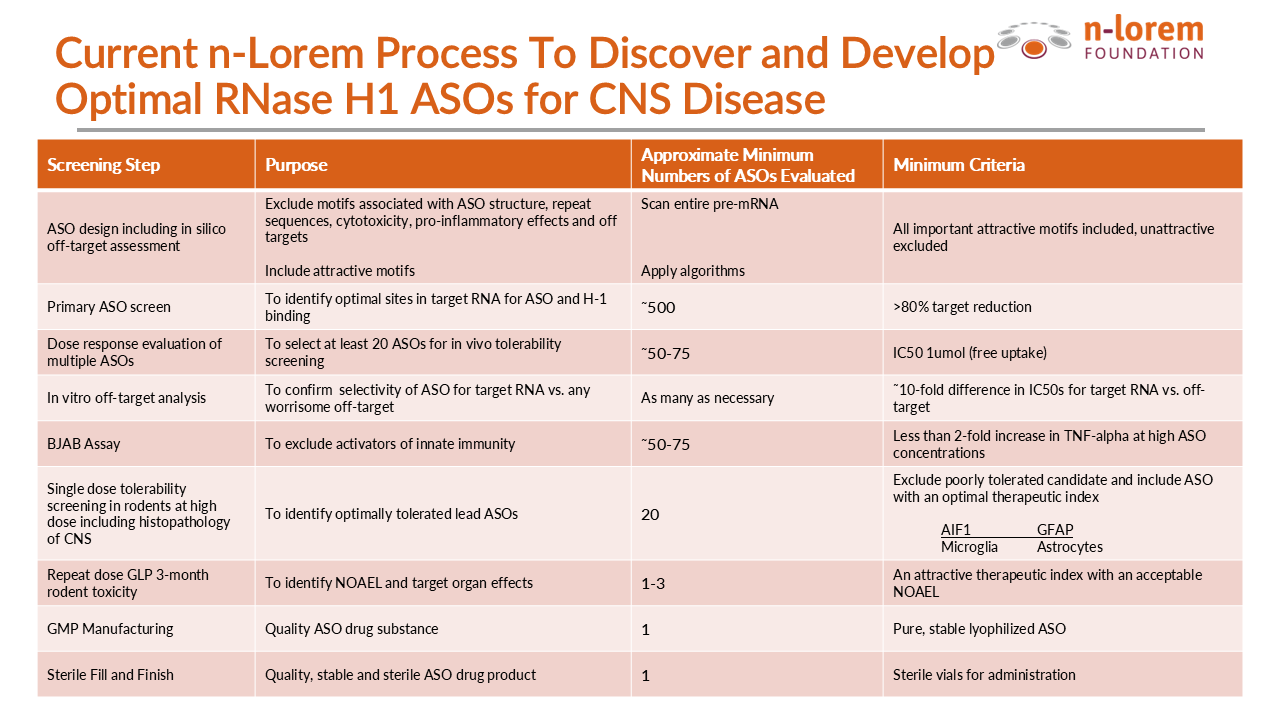
The table below shows the screening process we currently use at n-Lorem to identify optimal RNase H1 gapmer PS ASOs for CNS targets.
The n-Lorem ASO design algorithm
The quality and value of any algorithm is determined by the quality and volume of data used to develop the algorithm. The algorithm at n-Lorem is based on 35 years of ASO experience, millions of PS ASOs of various chemical and mechanistic classes studied in many cell types, animal models and hundreds of thousands of humans, and it’s continuously informed by multiple published and in-house ‘omics’ datasets.
The algorithm is designed to prevent synthesis of ASOs that might bind to sequences such as poly-pyrimidine tracts, that are present in all pre- and m-RNAs and to exclude certain PS ASO sequence motifs such as CpGs that are known to be problematic or form structures in PS ASOs. It is a “learning program” that evolves as we accumulate new data and deeper understanding of the molecular mechanisms through which PS ASOs cause the effects that we observed. There is no other algorithm that benefits from the cumulative experience with ASOs at previously Ionis and now n-Lorem.
In vitro screening to identify potent PS ASOs
To begin with, we enter the entire sequence of the target RNA into our design algorithm which then designs about 500 different PS ASOs to bind to the corresponding number of different sites in the target RNA. We first screen all PS ASOs at a single concentration to identify potentially potent and attractive ASOs. This usually identifies 50 or more ASOs that may be attractive. Those ASOs are then subjected to a study that evaluates the effect of 5 different ASO concentrations on the target RNA. This gives us a very accurate estimate of potency and maximum target RNA reduction.
Why do we study a minimum of 500 or so ASOs? Because long ago, we evaluated the benefits of studying from 100 to 100,000 different ASOs per RNA target and found that about 500 was sufficient to make it highly likely that a very potent ASO would be identified. Why do we study 5 different concentrations of each lead ASO? Because 5 concentrations assures that we have accurate estimates of potency and maximum RNA target reduction.
We typically conduct all the studies described above in patient derived cells. Often, and where applicable, we confirm the results in patient derived cells in cell lines maintained in the laboratory that express the target RNA. We always repeat these studies multiple times to be certain that all the results are reproducible.
Evaluation of selectivity for the target RNA
To exclude the possibility that we picked a lead ASO designed to bind a sequence in its target RNA could also bind the same sequence in other RNAs, which also play important roles in the patient’s cells, we first perform an in silico (computer only) analysis of all cellular RNAs to see if there are worrisome sequences in other non-targeted RNAs. RNA binding theory and our experience teach that sequence mismatches (a nucleotide in the RNA that does not bind to the nucleotide in the ASO, for example an A across from a G in the ASO) in the center of an ASO binding site is more disruptive of ASO binding and RNase H1 cleavage than a mismatch at either end of the ASO binding site. We also know that even two mismatches in a 20mer ASO are typically sufficient to assure that no effects on that non-target RNA are likely.
If we identify RNAs in which there is a sequence that might interact with the ASO and the non-target is expressed in the CNS (for CNS indications), we then evaluate that possibility by looking at that non-target RNA in lab experiments. If the lead ASO has a meaningful effect on an important non-target RNA, we discard that ASO.
Potential cytotoxicity
Some PS ASOs can be directly toxic to cells. Fortunately, thanks to work from my lab, we understand the mechanisms of cytotoxicity and have very specific markers that we can evaluate to exclude PS ASOs that are toxic to the cell.
Potential immunotoxicity
All cells have systems to identify foreign nucleic acids like PS ASOs and this system is called the innate immune system. As a product of years of research, we identified a particular cell line called the BJAB cell line that is very good at identifying PS ASOs that could cause an immunological side effect and all lead ASO are studied at high concentrations in that cell line.
CNS toxicity screening in rats
Based on decades of experience, we know that intrathecal dosing of lead PS ASOs in rats identifies potentially problematic PS ASOs. Consequently, we evaluate the effects of about 20 lead PS ASOs at a high dose in rats after intrathecal administration (injection into rat spinal fluid). Why 20 lead ASOs? Because experience has shown that 20 lead PS ASOs is the minimum number needed to make it likely that we will find one or more well tolerated PS ASOs. Based on our experience, we have sophisticated scoring systems that we use to identify well tolerated PS ASOs for the CNS.
We know that a certain type of cell in the CNS, Purkinje cells, is particularly sensitive to direct PS ASO toxicity, so we pay very close attention to them, and we have identified markers of activation of the immune cells in the CNS, and we evaluate them.
GLP rat toxicology study
The best PS ASO is then evaluated at 2 dose levels for three months in the rat. These studies are large, well-controlled and required by the FDA. We once again evaluate clinical signs, neurobehavioral parameters, other markers of inflammation and examine the CNS tissues after dosing using standard histopathology methods.
Conclusions
The n-Lorem ASO design and screening process derives from decades of experience, is multistep and involves hundreds of PS ASOs for each target. We are confident that this process, and the judgment that comes from decades of experience, are the reasons we have achieved such an excellent clinical safety and tolerability record.

We cannot do
this alone
Together we are changing the world—
one patient at a time
We hope that you join us on this journey to discover, develop and provide individualized antisense medicines for free for life for nano-rare patients. The ultimate personalized medicine approach – for free, for life.
Follow us on social for updates on our latest efforts


